Background of the cell states gradients
In the BPTT of LSTM, each loss gradient with respect to the weights shared at all steps is a summation of both short-term and long-term temporal-dependent gradients. In theory LSTM prevents vanishing gradients (equivalently captures long-term dependency) by keeping those long-term gradients away from zero. See blog for more details: Why LSTMs Stop Your Gradients From Vanishing: A View from the Backwards Pass
In particular Noh (2021) plotted dc_t/dc_0 over the steps t as an indicative statistic for the extent of vanishing gradients in LSTM. This quantity stands for the loss gradient over the longest temporal span at each step t. By collecting this quantity and plot it against the steps we can hopefully see the extent of vanishing gradients clearly.
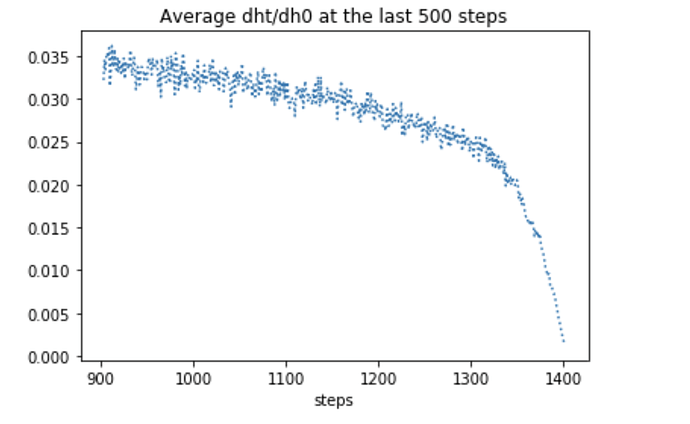
Note that in standard RNNs we need to plot the quantity dh_t/dh_0, where h_0 is the initial hidden states. For instance I have plotted the behaviour of dh_t/dh_0 for the t = 1,…,500 and t = 900,…,1400.
Now the task for LSTM:
I have experimented on a text comment dataset where each comment is labeled with a binary toxicity index. I have prepared my training sample by:
-
Take all 20000 toxic comments and another 20000 non-toxic comments from the data.
The idea is to increase the concentration of toxic inputs (to 50%). -
Replace the last 30% of words in each comment with the word ‘the’.
Comments that are labeled ‘toxic’ generally contain toxic content throughout. The idea is to remove any toxicity in the last bit of each comment in order to test the ability of the model in ‘memorising’ the toxic content in earlier steps.
tf.gradient is an expensive method, so I have computed dc_t/dc_0 only for the first and last 20 steps, but found the partial derivatives are all zeros. I would like some help for understanding this behaviour.
Imports, preprocessing and model set-up:
import numpy as np
from numpy import array, asarray, zeros
import pandas as pd
from tqdm import tqdm
from sklearn.model_selection import train_test_split
import tensorflow as tf
from keras import Input, Model
from keras.models import Sequential
from keras.layers.recurrent import LSTM, GRU, SimpleRNN
from keras.layers.core import Dense, Activation, Dropout, Flatten
from keras.layers.embeddings import Embedding
from sklearn import preprocessing, decomposition, model_selection, metrics, pipeline
from keras.layers import RNN, LSTMCell, Flatten, Bidirectional, SpatialDropout1D
from keras.preprocessing import sequence, text
from keras.callbacks import EarlyStopping, LambdaCallback, TensorBoard
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
from keras import backend as k
import os
### 2. Pre-processing
train= pd.read_csv('/Users/jigsaw-toxic-comment-train.csv')
train.drop(['severe_toxic','obscene','threat','insult','identity_hate'],axis=1,inplace=True)
sentences_t= train[train['toxic']== 1]
sentences_nt= train[train['toxic']== 0]
sentences_t= sentences_t[:20000]
sentences_nt= sentences_nt[:20000]
sentences_half= pd.concat([sentences_t, sentences_nt])
# Shuffle the half-half data:
sentences_half = sentences_half.sample(frac=1).reset_index(drop=True)
# Replace last 30% of each sentence with irrelevant words:
idx= 0.3
sentences_mod= sentences_half.copy()
for i in range(len(sentences_mod)):
sentence= sentences_mod['comment_text'].iloc[i]
split_sent= str(sentence).split()
sent_length= len(split_sent)
n_rep= int(np.floor(sent_length* idx))
split_sent[-n_rep:]= ['the']*n_rep
sentences_mod['comment_text'].iloc[i]= ' '.join(word for word in split_sent)
# Tokenisation, Prepare GloVe 6B 100d embeddings:
xm= sentences_mod['comment_text']; ym= sentences_mod['toxic']
tok_m= text.Tokenizer()
tok_m.fit_on_texts(list(xm))
input_dim_m= len(tok_m.word_index)+1
input_length_m= sentences_half['comment_text'].apply(lambda x: len(str(x).split())).max()
xm_seq= tok_m.texts_to_sequences(xm)
longmod_pad= sequence.pad_sequences(xm_seq, maxlen= input_length_m)
print('Shape of long modified sentences input object:', longmod_pad.shape)
print('Max len of long modified sentences:', input_length_m)
output_dim= 100
def embed_mat(input_dim, output_dim, tok):
'''By default output_dim = 100 for GloVe 100d embeddings'''
embedding_dict=dict()
f= open('/Users/glove.6B.100d.txt')
for line in f:
values= line.split()
word= values[0]; coefs= asarray(values[1:], dtype= 'float32')
embedding_dict[word]= coefs
f.close()
Emat= zeros((input_dim, output_dim))
for word, i in tok.word_index.items():
embedding_vector= embedding_dict.get(word)
if embedding_vector is not None:
Emat[i]= embedding_vector
print('Embedding weight matrix has shape:', Emat.shape)
return Emat
Emat_m= embed_mat(input_dim_m, output_dim, tok_m)
### 3. LSTM model that outputs cell states only:
batch_size = 100
inp= Input(batch_shape= (batch_size, input_length_m), name= 'input')
emb_out= Embedding(input_dim_m, output_dim, input_length= input_length_m,
weights= [Emat_m], trainable= False, name= 'embedding')(inp)
class LSTMCellwithStates(LSTMCell):
def call(self, inputs, states, training=None):
real_inputs = inputs[:,:self.units] # decouple [h, c]
outputs, [h,c] = super().call(real_inputs, states, training=training)
return tf.concat([h, c], axis=1), [h,c]
rnn = RNN(LSTMCellwithStates(200), return_sequences= True, return_state= False, name= 'LSTM')
h0 = tf.Variable(tf.random.uniform((batch_size, 200)))
c0 = tf.Variable(tf.random.uniform((batch_size, 200)))
rnn_allstates= rnn(emb_out, initial_state=[h0, c0])
model_lstm_mod = Model(inputs=inp, outputs= rnn_allstates[:, :, 200:], name= 'model_LSTMCell')
model_lstm_mod.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['acc'])
model_lstm_mod.summary()
Obtaining the partial derivatives:
Note that I am not doing for epochs, but only over some batches because it is very slow.
nrows= 400
ds = tf.data.Dataset.from_tensor_slices((longmod_pad[:nrows], ym[:nrows])).batch(100)
@tf.function
# Compute gradients
def compute_dct_dc0(t, x, c0):
return tf.gradients(model_lstm_mod(x)[:,t,:], c0)
n_b = int(nrows/ 100) # 10 batches
n_steps = 20 # look up only the first and last 20 steps
dctdc0_last20= tf.zeros([n_b, n_steps])
for b, (x_batch_train, y_batch_train) in enumerate(ds): # batches 0,1
grad_batch= [] # a list of 1403 scalar gradients on the current batch
for t in range(input_length_m - n_steps, input_length_m):
dctdc0_b_t = compute_dct_dc0(t, x_batch_train, c0) # (batch_size, n_units)
grad_t = tf.reduce_mean(abs(dctdc0_b_t[0]), [0,1]) # Scalar dctdc0 at the current batch and step
print('step', t+1, 'of batch' ,b+1, 'done')
grad_batch.append(grad_t)
dctdc0_last20= tf.concat([dctdc0_last20, [grad_batch]], axis = 0)
dctdc0_last20_agg= tf.reduce_mean(dctdc0_last20, 0) # take rowmean to obtain a vector of shape (20,)
In particular these gradients are exact zeros, not just close to zero. That is a strong indication of vanishing gradients.
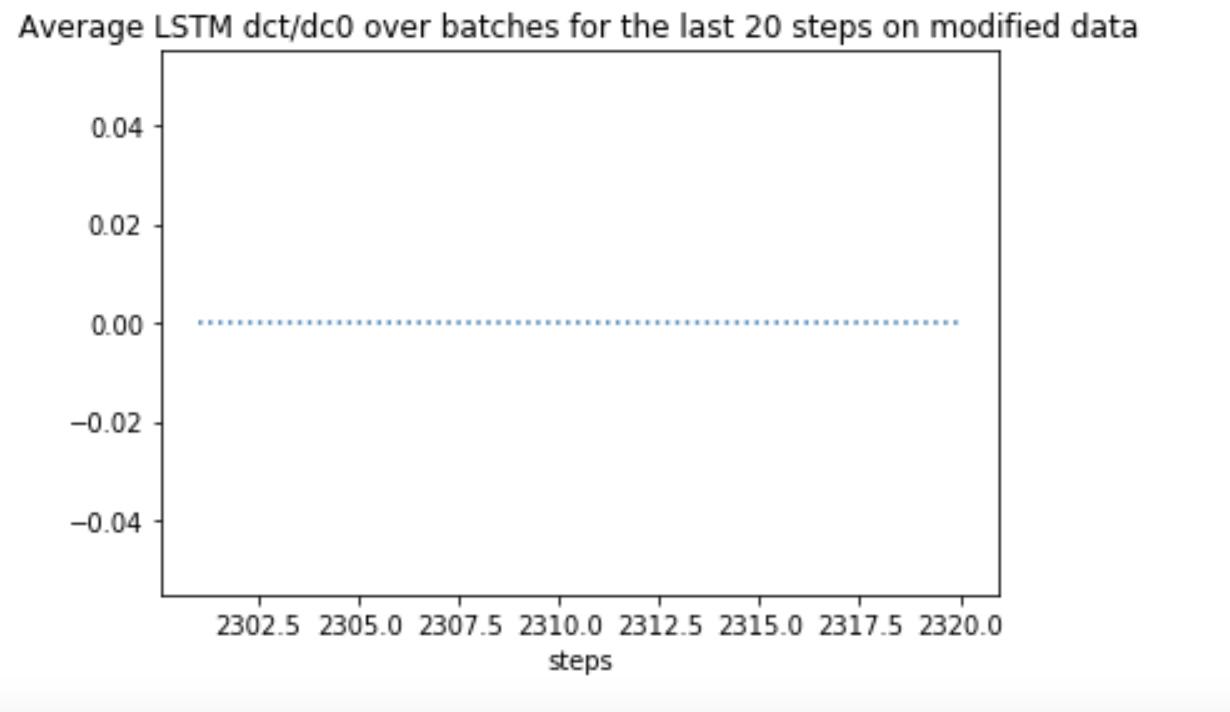
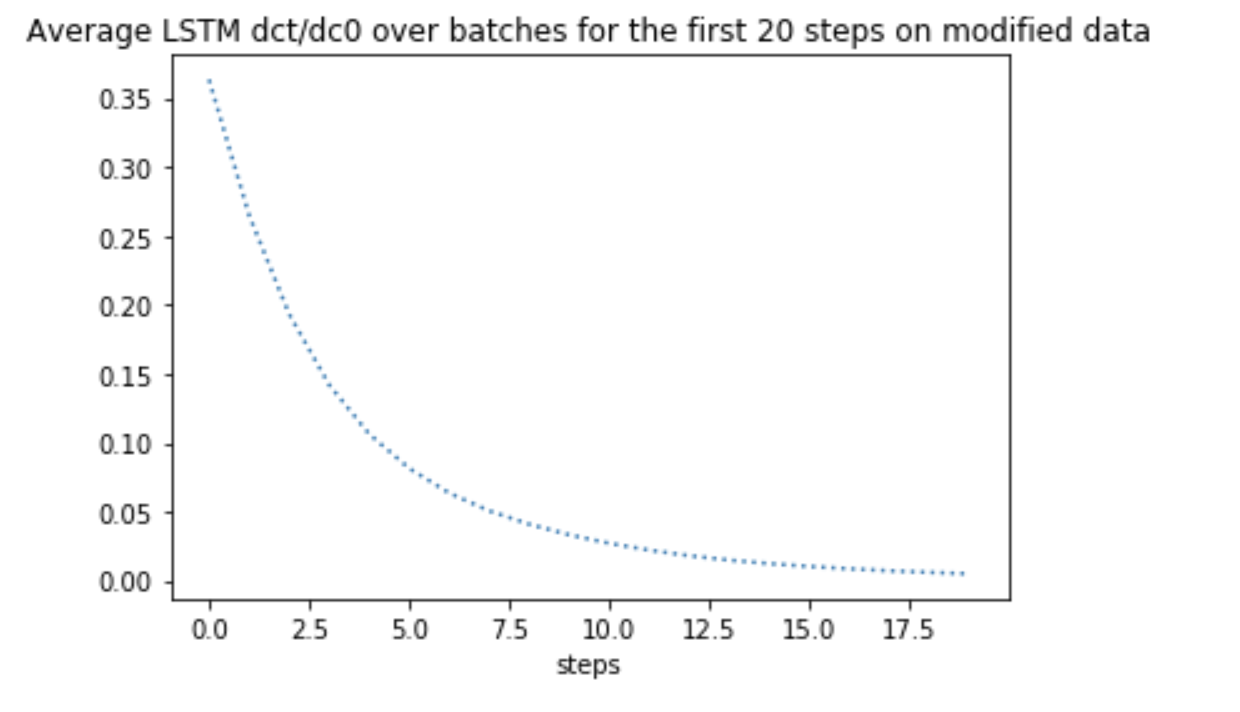
Same gradients on the first 20 steps showed a sharp decrease in mean absolute value. Why LSTM is not preventing vanishing gradients?
Minimial Example:
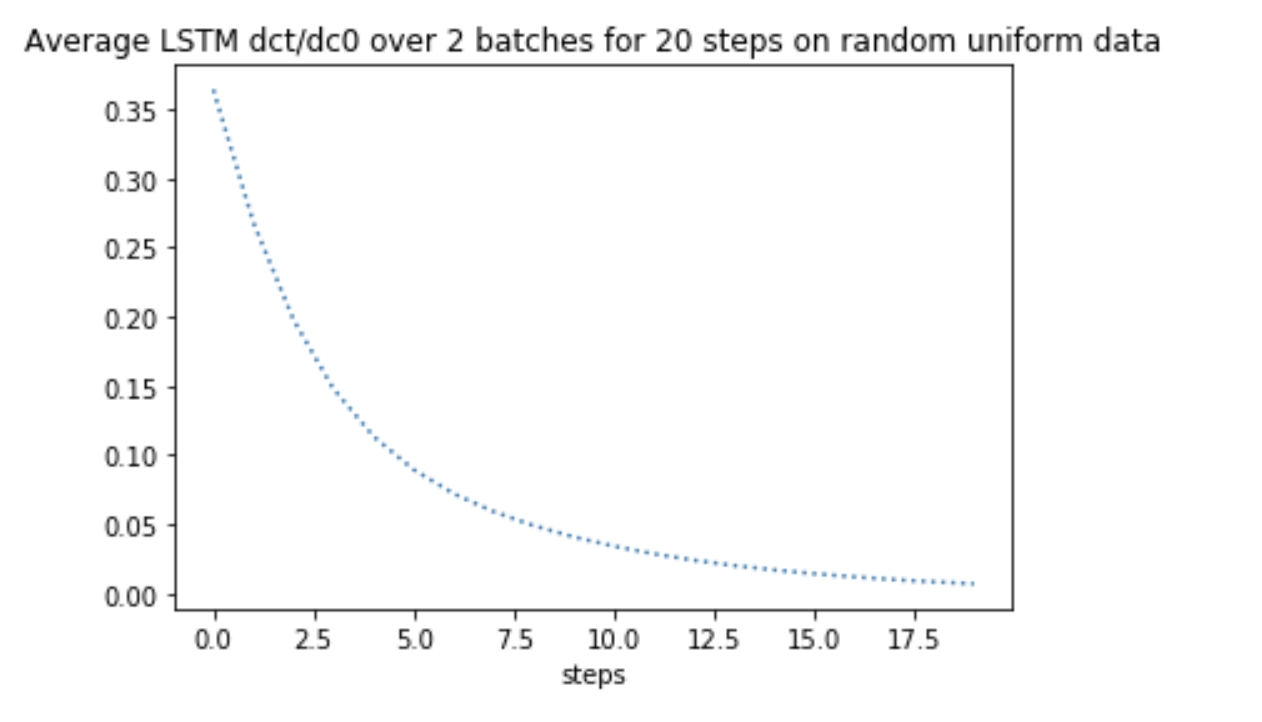
As a reproducible example, I have used random uniform data to carry out the same computations. I found that the gradients dc_t/dc_0 behaves EXACTLY the same as it behaves on the text comment data!
It seems that my code is not capturing some critical information in computation, thus leading to the same behaviour on different datasets. Where have I gone wrong?
# For each c_t, compute gradient dc_t/dc_0:
batch_size = 100; input_length_m = 20
xtr_pad = tf.random.uniform((batch_size*2, input_length_m), maxval = 500, dtype=tf.int32)
ytr = tf.random.normal((batch_size*2, input_length_m, 200))
inp= Input(batch_shape= (batch_size, input_length_m), name= 'input')
emb_out= Embedding(500, 100, input_length= input_length_m, trainable= False, name= 'embedding')(inp)
class LSTMCellwithStates(LSTMCell):
def call(self, inputs, states, training=None):
real_inputs = inputs[:,:self.units] # decouple [h, c]
outputs, [h,c] = super().call(real_inputs, states, training=training)
return tf.concat([h, c], axis=1), [h,c]
rnn = RNN(LSTMCellwithStates(200), return_sequences= True, return_state= False, name= 'LSTM')
h0 = tf.Variable(tf.random.uniform((batch_size, 200)))
c0 = tf.Variable(tf.random.uniform((batch_size, 200)))
rnn_allstates= rnn(emb_out, initial_state=[h0, c0])
model_lstm_mod = Model(inputs=inp, outputs= rnn_allstates[:, :, 200:], name= 'model_LSTMCell')
model_lstm_mod.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['acc'])
### Compute gradients:
ds = tf.data.Dataset.from_tensor_slices((xtr_pad, ytr)).batch(100)
@tf.function
# Compute gradients
def compute_dct_dc0(t, x, c0):
return tf.gradients(model_lstm_mod(x)[:,t,:], c0)
n_b = int(xtr_pad.shape[0]/ 100) # 200 batches
n_steps = 20 # look up only the first and last 20 steps
dctdc0_all= tf.zeros([n_b, n_steps])
for b, (x_batch_train, y_batch_train) in enumerate(ds): # batches 0,1
grad_batch= [] # a list of 1403 scalar gradients on the current batch
for t in range(n_steps):
# steps 0,...,19
dctdc0_b_t = compute_dct_dc0(t, x_batch_train, c0) # (batch_size, n_units)
grad_t = tf.reduce_mean(abs(dctdc0_b_t[0]), [0,1]) # Scalar dctdc0 at the current batch and step
print('step', t+1, 'of batch' ,b+1, 'done')
grad_batch.append(grad_t)
dctdc0_all= tf.concat([dctdc0_all, [grad_batch]], axis = 0)
dctdc0_agg= tf.reduce_mean(dctdc0_all, 0) # take rowmean to obtain a vector of shape (20,)
print(dctdc0_agg.shape)
Loss and accuracy for the same task:
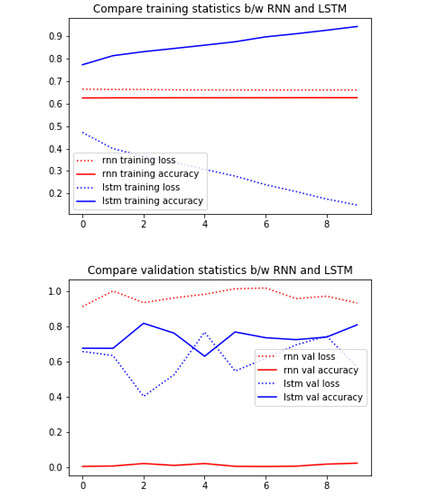
I have compared the loss and accuracy for 10 epochs between RNN and LSTM on the same task, and found that LSTM exhibits superior performance, as we should expect.
Therefore it is really abnormal to have hidden states gradients of RNN behaving better than the cell states gradients of LSTM.
Thanks in advance for any idea on this long question : )