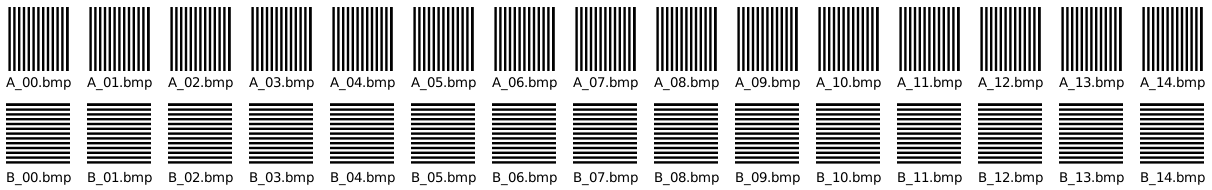
I want to build a Siamese network for biometric fingerprint matching. For first tests, I have generated a tiny artificial dataset: two “fingers” A and B with vertical resp. horizontal stripes. (15 images of each, corresponding to 15 scans of each finger.)
Dataset creation
from PIL import Image, ImageDraw
import os
width = 160
height = 160
img = Image.new('L', (width, height), 'white')
stripe_width = 6
num_stripes = int(width / stripe_width)
for i in range(num_stripes):
color = 0 if i % 2 else 255
# Set the coordinates for the stripe
x0 = i * stripe_width
y0 = 0
x1 = x0 + stripe_width
y1 = height
img_draw = ImageDraw.Draw(img)
img_draw.rectangle([x0, y0, x1, y1], fill=color)
rotated = img.rotate(90)
path = 'data/pseudo_fingerprints/'
os.mkdir(path)
for i in range(15):
img.save(path + f'A_{i:02d}.bmp')
rotated.save(path + f'B_{i:02d}.bmp')
My code is an adaptation of a Kaggle notebook by Pere Martra for MNIST and looks like this:
Imports and data preparation
# Siamese network for image comparison.
#
# License: Apache 2.0
# Authors: Pere Martra, Robert Pollak
from tqdm import tqdm
import os
import glob
import matplotlib as mpl
import numpy as np
np.random.seed(42)
import tensorflow as tf
tf.random.set_seed(42)
from tensorflow.keras.models import Model
from tensorflow.keras.layers import Input, Flatten, Dense, Dropout, Lambda
from tensorflow.keras import backend as K
from tensorflow.keras import optimizers
# Use only as much GPU memory as needed.
# Source: https://www.tensorflow.org/guide/gpu#limiting_gpu_memory_growth
gpus = tf.config.list_physical_devices('GPU')
# Currently, memory growth needs to be the same across GPUs
for gpu in gpus:
tf.config.experimental.set_memory_growth(gpu, True)
#%% Prepare the data
data_path = 'data/pseudo_fingerprints'
image_edge = 160
all_image_names = sorted(
map(os.path.basename,
glob.iglob(os.path.join(data_path, '*.bmp'))))
# Filename example: A_09.bmp
get_finger = lambda name: name[:1]
fingers = sorted(set(map(get_finger, all_image_names)))
X_model = []
y_model = []
for i_finger, finger in tqdm(enumerate(fingers)):
image_names = sorted(
map(os.path.basename,
glob.iglob(os.path.join(data_path, finger + '*.bmp'))))
for image_name in image_names:
image = mpl.image.imread(os.path.join(data_path, image_name))
X_model.append(image)
y_model.append(i_finger)
X_model = np.array(X_model)
y_model = np.array(y_model)
# Add the channel dimension.
X_model = X_model.reshape((-1, image_edge, image_edge, 1))
print(f'{X_model.shape=}')
# Normalize the data.
X_model = X_model.astype('float32') / 255
# Mix the modeling images before partitioning.
shuffled = np.arange(len(y_model))
np.random.shuffle(shuffled)
X_model = X_model[shuffled]
y_model = y_model[shuffled]
# The parameter min_equals indicates how many equal pairs we want in the dataset at least.
# If we just created random pairs, the number of equal pairs would be very small.
def create_pairs(X, y, min_equals):
pairs = []
labels = []
equal_items = 0
# Indices of all the positions containing the same value.
index = [np.where(y == i)[0] for i in range(10)]
for n_item in range(len(X)):
if equal_items < min_equals:
# Select the number to pair from index containing equal values.
num_rnd = np.random.randint(len(index[y[n_item]]))
num_item_pair = index[y[n_item]][num_rnd]
equal_items += 1
else:
# Select any number in the list.
num_item_pair = np.random.randint(len(X))
# Matching images are labeled with 1, others with 0.
labels += [int(y[n_item] == y[num_item_pair])]
pairs += [[X[n_item], X[num_item_pair]]]
return np.array(pairs), np.array(labels).astype('float32')
#%%% Create training and validation dataset.
val_size = len(X_model) // 5
X_val = X_model[:val_size]
y_val = y_model[:val_size]
X_train = X_model[val_size:]
y_train = y_model[val_size:]
training_equals = len(X_train) // 3
training_pairs, training_labels = create_pairs(X_train, y_train, min_equals=training_equals)
val_equals = val_size // 3
val_pairs, val_labels = create_pairs(X_val, y_val, min_equals=val_equals)
Model creation
#%% Create the Siamese model
#%%% Common part
def initialize_base_branch():
input = Input(shape=(image_edge, image_edge,), name="base_input")
x = Flatten(name="flatten_input")(input)
x = Dense(128, activation='relu', name="first_base_dense")(x)
x = Dropout(0.3, name="first_dropout")(x)
x = Dense(128, activation='relu', name="second_base_dense")(x)
x = Dropout(0.3, name="second_dropout")(x)
x = Dense(128, activation='relu', name="third_base_dense")(x)
# Returning a Model, with input and outputs, not just a group of layers.
return Model(inputs=input, outputs=x)
base_model = initialize_base_branch()
#%%% Siamese part with two inputs
# Input for the left part of the pair. We are going to pass training_pairs[:,0] to this layer.
input_l = Input(shape=(image_edge, image_edge,), name='left_input')
#Attention: base_model is not a function, it is a model, and we are adding our input layer.
vect_output_l = base_model(input_l)
# Input layer for the right part of the siamese model. Will receive training_pairs[:,1].
input_r = Input(shape=(image_edge, image_edge,), name='right_input')
vect_output_r = base_model(input_r)
def euclidean_distance(vects):
x, y = vects
sum_square = K.sum(K.square(x - y), axis=1, keepdims=True)
return K.sqrt(K.maximum(sum_square, K.epsilon()))
def eucl_dist_output_shape(shapes):
shape1, shape2 = shapes
return (shape1[0], 1)
# The lambda output layer calling the euclidean distances, will return the difference between both vectors.
output = Lambda(euclidean_distance, name='output_layer',
output_shape=eucl_dist_output_shape)([vect_output_l, vect_output_r])
# The overall model has two inputs and one output. Each of the inputs contains the common model.
model = Model([input_l, input_r], output)
Training and evaluation
#%% Train
# With a big margin, the dissimilarities have more weight than the similarities.
def contrastive_loss_with_margin(margin):
def contrastive_loss(y_true, y_pred):
square_pred = K.square(y_pred)
margin_square = K.square(K.maximum(margin - y_pred, 0))
return (y_true * square_pred + (1 - y_true) * margin_square)
return contrastive_loss
opt = optimizers.RMSprop()
model.compile(loss=contrastive_loss_with_margin(margin=1), optimizer=opt)
history = model.fit(
[training_pairs[:,0], training_pairs[:,1]],
training_labels, epochs=500,
batch_size=128,
validation_data = ([val_pairs[:, 0], val_pairs[:, 1]], val_labels),
)
#%% Evaluate
# Assuming that with a difference less than 0.5 the image pair matches.
def compute_accuracy(y_true, y_pred):
pred = y_pred.ravel() < 0.5
return np.mean(pred == y_true)
y_pred_train = model.predict([training_pairs[:,0], training_pairs[:,1]])
train_accuracy = compute_accuracy(training_labels, y_pred_train)
y_pred_val = model.predict([val_pairs[:,0], val_pairs[:,1]])
val_accuracy = compute_accuracy(val_labels, y_pred_val)
print(f'{train_accuracy=:.4f}, {val_accuracy=:.4f}')
Unfortunately, the training loss does not get lower than about 0.25, and the training accuracy is only 0.54. A lower learning rate also does not help. Why doesn’t this modeling work?
(I am using TensorFlow 2.9.x.)