Following the image classification tutorial using google colab and my own data. The only adjustment i make to the code is the following
import pathlib
dataset_url = "https://storage.googleapis.com/download.tensorflow.org/example_images/flower_photos.tgz"
data_dir = tf.keras.utils.get_file('flower_photos.tar', origin=dataset_url, extract=True)
data_dir = pathlib.Path(data_dir).with_suffix('')
data_dir = pathlib.Path('/root/.keras/datasets/multiple_pics')
(1) Data
I have taken images of lego blocks. There are two types:
labeled using directory folder structure as
/images
/rect_2
/rect_8
Here are two example images from each directory respectively:


Now, the results are wildy innacurate but the accuracy is often a minimimum of 99 percent but many times 100 percent. But it will call 2 x1 lego block (labeled as rect_2) rect_8 with 100 percent confidence with far too much failure.
The training and validation plots look as follows for (1) the non-augmented and (2) the augmented.
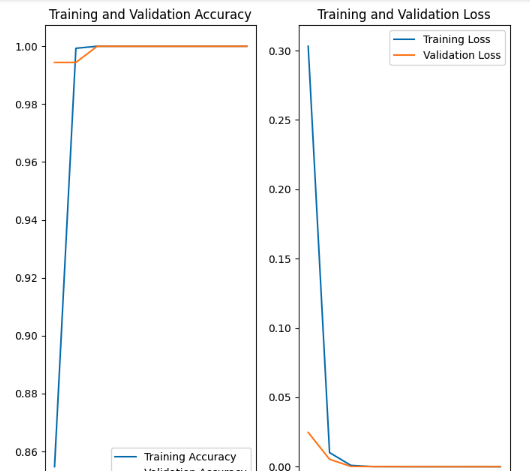
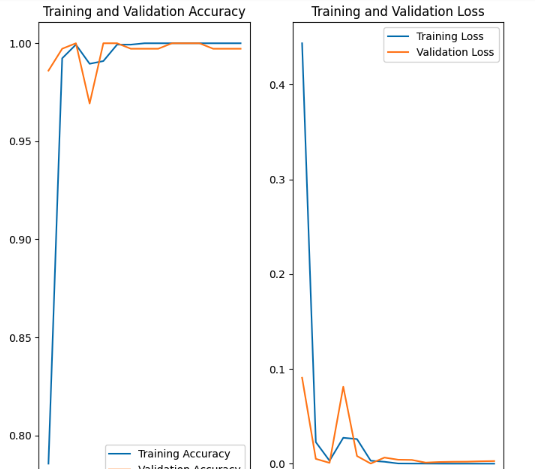
Finally the augmented images look good. Here is an example of the rect_8 or 4x2 lego block. I am at a complete loss having little experience with this technology but many hours trying to adjust the data accordingly.
Any help would be appreciated.
AUGMENTED IMAGES

EXAMPLE OF THE PLOT OF THE LABELS AND THEIR IMAGES
